 Media 000669696: Fossil Skull Cap
Restricted Download
Type
Volumetric Image Series
Modality
X-Ray Computed Tomography (CT/microCT)
Media represents biological specimen
Managed by
Media 000669696: Fossil Skull Cap
Restricted Download
Type
Volumetric Image Series
Modality
X-Ray Computed Tomography (CT/microCT)
Media represents biological specimen
Managed by
Media 000669696: Fossil Skull Cap
Restricted Download
Type
Volumetric Image Series
Modality
X-Ray Computed Tomography (CT/microCT)
Media represents biological specimen
Managed by
Login or create an account to download files
GENERAL DETAILS Field Definitions
Media ID
000669696
Media type
Volumetric Image Series
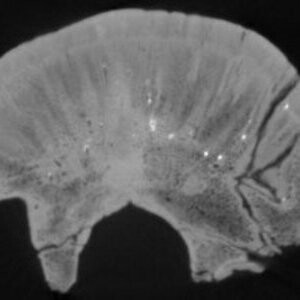
Object element or part
fossil skull cap
Object represented
Object taxonomy
Stegoceras validum
Object organization
Side
--
Orientation
--
Short description
HRXCT scan of fossil skull cap
Full description
University of Texas High-Resolution X-ray CT Facility Archive 2290
Evans:
stegD: Scans of the fossil skull cap of Stegoceras validum (TMP 99.62.1; Ornithischia, Pachycephalosauridae; Upper Cretaceous; Belly River Group, Dinosaur Park Fm; Alberta, Irvine) for David Evans of the Royal Ontario Museum. Specimen scanned by Jessie Maisano 25 June 2010.
16bitrot: 1024x1024 16-bit TIFF images. II, 200 kV, 0.18 mA, intensity control off, high-power mode, no filter, empty container wedge, no offset, slice thickness 1 line ( = 0.08602 mm), S.O.D. 247 mm, 1400 views, 2 samples per view, inter-slice spacing 1 line ( = 0.08602 mm), field of reconstruction 82 mm (maximum field of view 82.16 mm), reconstruction offset 3000, scanner scale 5400, reconstruction scale 7000. Acquired with 31 slices per rotation and 25 slices per set. Fillin-, streak-, drift- and ring-removal processing done by Satomi Ishihama based on correction of raw sinogram data using IDL routines “RK_SinoFillin” with parameters “100, /top” (for slices 838-868) and “115, /top” (for slices 1365-1395), “RK_SinoDeStreak” with parameter “cutoff=0.07,” “RK_SinoDeDrift” with default parameters and “RK_SinoRingProcSimul” with parameters “binwidth=21, bestof5=11.” Reconstructed with rotation angle=-24.1 degrees (for slices 844-1364), -29.2 degrees (for slices 1371-1612), and beam-hardening coefficients using IDL FindBHC [Y found=-0.06, 0.06, 0.16, 0.28, 0.41, 0.55, 0.71, 0.90, 1.15, 1.59 with parameters “Num Coef=10, Downsample=none, ROI mode=bi-joint”]. Deleted first 6 duplicate slices from each rotation. Rotation correction processing done by Satomi Ishihama using IDL routine “DoRotationCorrection.” Post-reconstruction ring correction applied by Satomi Ishihama using the parameters oversample = 3.0, binwidth = 17 or 21, sectors =1 or 60, smoothing = median or mean for selected slices over the gray scale range 1280-65535. Deleted first 7 and last 21 blank slices. Total final slices = 1272.
Creator
Jessie Maisano
Date created
June 25, 2010
Date uploaded
October 09, 2024
FILE OBJECT DETAILS Field Definitions
File name
16bitrot.zip
File format(s)
ZIP archive, TIFF
File size
1.29 GB
Image width
1019
Image height
671
Color space
BlackIsZero
Color depth
16
Compression
Uncompressed
Series type
Reconstructed image stack
X pixel spacing
0.080080
Y pixel spacing
0.080080
Z pixel spacing
0.086020
Pixel spacing units
Mm
Slice thickness
--
Number of images in set
1,272
IMAGE ACQUISITION AND PROCESSING AT A GLANCE Field Definitions
Number of parent media
0
Number of processing events
1
(0 steps)
Modality
X-Ray Computed Tomography (CT/microCT)
Device
Varian Medical Systems (Bio-Imaging Research, Inc) ACTIS CT scanner with FeinFocus X-ray source, The University of Texas High-Resolution X-ray Computed Tomography Facility (University of Texas at Austin)
OWNERSHIP AND PERMISSIONS Field Definitions
Data managed by
Data uploaded by
Publication status
Restricted Download
Download reviewer
IP holder
Royal Tyrrell Museum
Copyright statement
Creative Commons license
--
Morphosource use agreement type
Standard
Permits commercial use
Commercial Use Not Permitted
Permits 3D use
3D Printing Limited
Required archival of published derivatives
On MorphoSource
Funding attribution
DBI-1902242
Publisher
oUTCT PEN
Cite as
These data were made available by David Evans. Data upload to MorphoSource was funded by DBI-1902242. The files were downloaded from www.MorphoSource.org, Duke University.
Media preview mode
Interactive/Embeddable
Additional usage agreement
None
IDENTIFIERS AND EXTERNAL LINKS Field Definitions
MorphoSource ARK
MorphoSource DOI
Not assigned
External identifier
--
External media URL
--
IMAGE ACQUISITION AND PROCESSING IN DETAIL
STEP 1: IMAGE ACQUISITION
Physical object
tmp:99.62.1
was imaged to create
undeposited raw media.
IMAGING DETAILS Field Definitions
Modality
X-Ray Computed Tomography (CT/microCT)
Device
Device facility
The University of Texas High-Resolution X-ray Computed Tomography Facility (University of Texas at Austin)
Creator
Jessie Maisano
Event date
June 25, 2010
Software
--
Description
--
Description attachment
None
Reference attachment
None
Exposure time
--
Flux normalization
--
Pixel spacing calibration
--
Shading correction
--
Frame averaging
--
Projections
--
Voltage
--
Power
--
Amperage
--
Surrounding material
--
X-ray tube type
--
Target type
--
Detector type
--
Detector pixels X
--
Detector pixels size X
--
Detector pixels Y
--
Detector pixels size Y
--
Detector configuration
--
Source object distance
--
Source detector distance
--
Target material
--
Rotation number
--
Phase contrast
--
Optical magnification
--
Acquisition type
--
STEP 2: IMAGE PROCESSING
Raw media (undeposited)
was processed
to create
derived
Media 000669696: Fossil Skull Cap [CTImageSeries] [CT] (this media).
Processing Details Field Definitions
Creator
--
Event date
--
Software
--
Description
--
Description attachment
None