 Media 000706839: Bony Labyrinth
Open Download
Type
Mesh
Modality
X-Ray Computed Tomography (CT/microCT)
Media represents biological specimen
Managed by
Media 000706839: Bony Labyrinth
Open Download
Type
Mesh
Modality
X-Ray Computed Tomography (CT/microCT)
Media represents biological specimen
Managed by
Media 000706839: Bony Labyrinth
Open Download
Type
Mesh
Modality
X-Ray Computed Tomography (CT/microCT)
Media represents biological specimen
Managed by
Login or create an account to download files
Media Display
In Collections
TAGS
GENERAL DETAILS Field Definitions
Media ID
000706839
Media type
Mesh
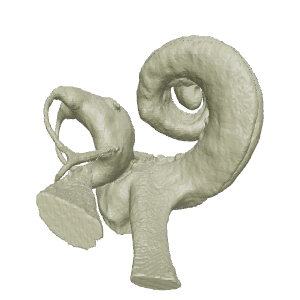
Object element or part
bony labyrinth
Object represented
Object taxonomy
Echovenator sandersi
Object organization
Side
--
Orientation
--
Short description
--
Full description
--
Creator
Rachel Racicot
Date created
February 04, 2025
Date uploaded
February 04, 2025
FILE OBJECT DETAILS Field Definitions
File name
Echovenator.stl
File format(s)
STL
File size
3.56 MB
Points
37289
Polygons
74588
Map type
--
UV coordinates
False
Vertex color
False
Bounding box dimensions
11.385, 9.374, 11.444
Centroid coordinates
1.643, -3.158, -6.096 (Bounding Box)
Units of point coordinates
--
IMAGE ACQUISITION AND PROCESSING AT A GLANCE Field Definitions
Number of parent media
1
Number of processing events
2
(1 step)
Derived directly from
Modality
X-Ray Computed Tomography (CT/microCT)
Device
Varian Medical Systems (Bio-Imaging Research, Inc) ACTIS CT scanner with FeinFocus X-ray source, The University of Texas High-Resolution X-ray Computed Tomography Facility (University of Texas at Austin)
OWNERSHIP AND PERMISSIONS Field Definitions
Data managed by
Data uploaded by
Publication status
Open Download
Download reviewer
IP holder
--
Copyright statement
--
Creative Commons license
--
Morphosource use agreement type
Standard
Permits commercial use
Commercial Use Not Permitted
Permits 3D use
3D Printing Limited
Required archival of published derivatives
On MorphoSource
Funding attribution
--
Publisher
--
Cite as
If data or derivatives are used please cite: Racicot RA, Mourlam MJ, Edkale EG, Glass A, Marino L, and Uhen M. Variation in whale (Cetacea) inner ear anatomy reveals the early evolution of “specialized” high-frequency hearing sensitivity. Journal of Anatomy. https://doi.org/10.1111/joa.14176
Media preview mode
Interactive/Embeddable
Additional usage agreement
None
IDENTIFIERS AND EXTERNAL LINKS Field Definitions
MorphoSource ARK
MorphoSource DOI
Not assigned
External identifier
--
External media URL
--
IMAGE ACQUISITION AND PROCESSING IN DETAIL
STEP 1: IMAGE ACQUISITION
Physical object
GSM:1098
was imaged to create
undeposited raw media.
IMAGING DETAILS Field Definitions
Modality
X-Ray Computed Tomography (CT/microCT)
Device
Device facility
The University of Texas High-Resolution X-ray Computed Tomography Facility (University of Texas at Austin)
Creator
--
Event date
--
Software
--
Description
Xenorophidae: Scans of the left petrosal of Xenorophidae (GSM 1098; Chandler Bridge; 27 MYA, Oligocene) for Dr. Mark Uhen of the Cranbrook Institute of Science and Dr. Lori Marino of the Department of Neuroscience and Behavioral Biology, Emory University. Specimen scanned by Matthew Colbert 10 November 2003.
16bit: 1024x1024 16-bit TIFF images. II, 180 kV, 0.133 mA, no filter, air wedge, no offset, slice thickness 2 lines (= 0.0622 mm), S.O.D. 90 mm, 1000 views, 2 samples per view, inter-slice spacing 2 lines (= 0.0622 mm), field of reconstruction 29 mm (maximum field of view 29.46449 mm), reconstruction offset 4200, reconstruction scale 1000. Acquired with 15 slices per rotation. Ring-removal processing done by Rachel Racicot based on correction of raw sinogram data using IDL routine “RK_SinoRingProcSimul”, with default parameters. Reconstructed with beam-hardening coefficients: 0.0, 0.9, 0.05. Total slices = 600.
8bitjpg: 8-bit JPG version of the above images.
Description Attachment
None
Photographic Reference
None
Filter
--
Exposure time
--
Flux normalization
--
Pixel spacing calibration
--
Shading correction
--
Frame averaging
--
Projections
--
Voltage
180 kV
Power
--
Amperage
0.133 mA
Surrounding material
--
X-ray tube type
--
Target type
--
Detector type
--
Detector pixels X
--
Detector pixels size X
--
Detector pixels Y
--
Detector pixels size Y
--
Detector configuration
--
Source object distance
--
Source detector distance
--
Target material
--
Rotation number
--
Phase contrast
--
Optical magnification
--
Acquisition type
--
STEP 2: IMAGE PROCESSING
Raw media (undeposited)
was processed
with 1 activity
to create
derived
Media 000703699: Left Periotic Of Echovenator Sandersi [Ct Image Series].
Processing Details Field Definitions
Creator
UTCT
Event date
November 10, 2003
Software
--
Description
Xenorophidae: Scans of the left petrosal of Xenorophidae (GSM 1098; Chandler Bridge; 27 MYA, Oligocene) for Dr. Mark Uhen of the Cranbrook Institute of Science and Dr. Lori Marino of the Department of Neuroscience and Behavioral Biology, Emory University. Specimen scanned by Matthew Colbert 10 November 2003.
16bit: 1024x1024 16-bit TIFF images. II, 180 kV, 0.133 mA, no filter, air wedge, no offset, slice thickness 2 lines (= 0.0622 mm), S.O.D. 90 mm, 1000 views, 2 samples per view, inter-slice spacing 2 lines (= 0.0622 mm), field of reconstruction 29 mm (maximum field of view 29.46449 mm), reconstruction offset 4200, reconstruction scale 1000. Acquired with 15 slices per rotation. Ring-removal processing done by Rachel Racicot based on correction of raw sinogram data using IDL routine “RK_SinoRingProcSimul”, with default parameters. Reconstructed with beam-hardening coefficients: 0.0, 0.9, 0.05. Total slices = 600.
8bitjpg: 8-bit JPG version of the above images.
Description attachment
None
Processing Activity 1: Reconstruction
Software
Description
Media produced by this event on MorphoSource
STEP 3: IMAGE PROCESSING
Derived
Media 000703699: Left Periotic Of Echovenator Sandersi [Ct Image Series]
was processed
to create
derived
Media 000706839: Bony Labyrinth [Mesh] [CT] (this media).
Processing Details Field Definitions
Creator
Rachel Racicot
Event date
--
Software
--
Description
STL file generated from the digital endocast of the bony labyrinth of _Echovenator_sandersi_
Description attachment
None